& Corresponding Authors
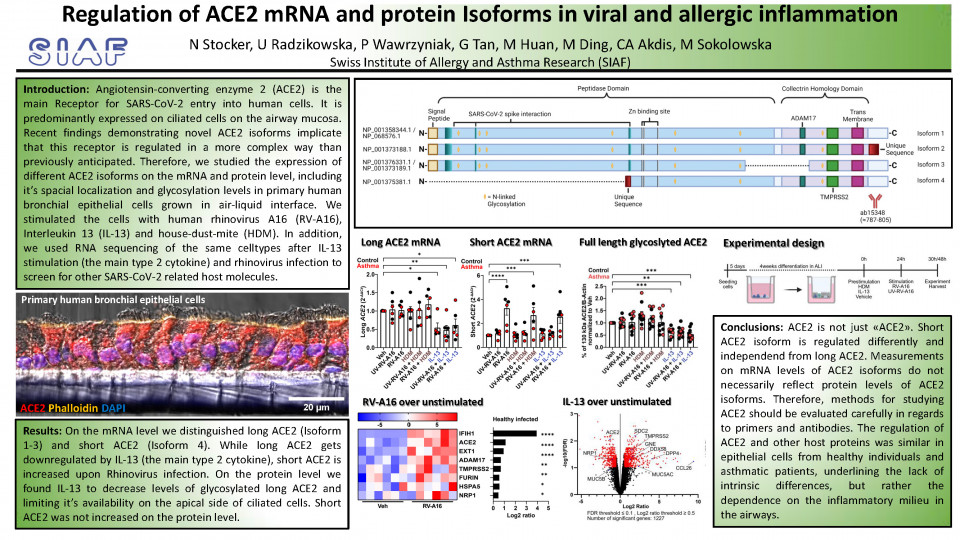
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infects airway epithelial cells via the main receptor Angiotensin-converting enzyme 2 (ACE2). ACE2 can be expressed in various isoforms (protein variants), which were found to be differentially regulated. One thereof called short ACE2 was found to be interferon regulated and does not possess the full N-terminal part, harboring the peptidase domain. Short ACE2 is not able to serve as a SARS-CoV-2 receptor, but was speculated to possess antiviral functions due to its interferon regulation. Expression of ACE2 mRNA by classical PCR and RNA-sequencing methods during the pandemics did not distinguish between different ACE2 isoforms on the mRNA level, leading to the contradictory results and especially concerns in interferon clinical trials in COVID-19 patients. The interaction between asthma, it´s viral or allergen-induced exacerbations and COVID-19 are still unclear. IL-13, the major type 2 asthma cytokine, was shown in mechanistic studies to inhibit SARS-CoV-2 infection in airway epithelium. Contrastingly, IL-13 has also been shown to be a major driver of COVID-19 severity and anti-IL4/13 treatment to have positive effects in COVID-19 clinical trials. Moreover, infection with rhinovirus was shown to inhibit SARS-CoV-2 infection by inducing interferons which act antiviral. On the other hand, we recently reported that co-infection of rhinovirus and SARS-CoV-2 leads to greater RIG-I inflammasome dependend damage in the airway epithelium in patients with asthma.
Therefore, since regulation of ACE2 isoforms in asthma has not been extensively studied, we investigated the impact of the major type 2 asthma cytokine (IL-13), house dust mite extract (HDM) exposure and viral infection by rhinovirus (RV-A16) on ACE2 in gene and protein expression. We used primary human bronchial epithelial cells from control and patients with asthma, grown and differentiated in air-liquid interface (ALI). These cells differentiate into basal (stem-cells), mucus-producing goblet- and ciliated cells and resemble the human bronchial epithelium as one of the most physiologically relevant model available to date.
By reverse transcription PCR (RT-PCR) we distinguished between long ACE2 and truncated (short) ACE2 mRNA transcripts and found that long ACE2 was downregulated by IL-13 treatment, whereas short ACE2 was upregulated by rhinovirus infection. ACE2 protein isoforms and posttranslational modifications (N-linked Glycosylation) were examined by western blotting, where we found IL-13 to reduce the amount of long glycosylated ACE2. Interestingly, we did not observe a significant change on short ACE2 protein level upon rhinovirus infection, as it was anticipated by observation on the RNA level. This suggests a high protein turnover with low protein stability, translation repression or mRNA degradation of this truncated isoform.
In view that IL-13 led to the decreased amount of long glycosylated ACE2, we continued to explore mechanisms which let to such alteration. By means of confocal microscopy on transversal cryosections of ALI cultures, we observed ACE2 to localize predominantly, but not restricted to the apical site of ciliated cells. IL-13 induced substantial morphological changes in the bronchial epithelium, which led to a redistribution of ACE2 signal. Apical ACE2 intensity, measured on top-view acquisition of ALI cultures, significantly dropped, which hints for less surface available ACE2. Additionally, we found IL-13 to induce changes in genes involved in ion- and transmembrane transport, lipid metabolic processes and protein glycosylation by using our bulk-RNA sequencing data of primary bronchial epithelial cells stimulated with IL-13.
We then screened for effects of IL-13 on other SARS-CoV-2 related host molecules in our bulk-RNA sequencing dataset and found besides others, TMPRSS2 and NRP1 to be differentially regulated. TMPRSS2 is a host protease which facilitates fusion of SARS-CoV-2 with the cellular membrane and NRP1 was shown to serve as an additional SARS-CoV-2 receptor. In a previously published dataset of bronchial epithelial cells infected with rhinovirus, we also found these two molecules to be differentially regulated. Therefore we also investigated the protein of TMPRSS2 and NRP1 by confocal microscopy and western blotting. We found TMPRSS2 and NRP1 both to be localizing apically on ciliated cells and nuclei, and remarkably strong signal of NRP1 in ciliae, making ciliated cells most susceptible for SARS-CoV-2 infection. By quantitative western blotting, we found alterations in their protein level to be negligible in any experimental condition. Overall, the regulation of ACE2 and other host factors was comparable in bronchial epithelial cells from control individual and patients with asthma.