Background
Allergies are a worldwide present affliction of dysregulated type 2 IgE-mediated immune responses against certain ‘harmless’ proteins from the environment. At the centre of allergic diseases stand B cells and their regulatory mechanisms. The regulation of allergen specific B cells in allergies, determines if an individual has an allergic or tolerant response to foreign antigens. The general agreement is, therefore, that in healthy and allergic individuals the B cell regulation is different. This means there should also be significant differences in B cell gene expression that would give insight to the underlying process of allergic dysregulation. This project aims to characterize B cells of allergic individuals on a transcriptomic level and investigate the differences of their gene expression in comparison to their healthy twins. The goal is to discover allergy relevant differential gene expression patterns and pathways between allergy discordant twins.
Methodology
We used bio banked PBMCs from 16 monozygotic twin pairs that are either allergy concordant or discordant. They were mainly allergic to pollen (birch and timothy grass) and/or house dust mites, while not undergoing any immunotreatment. We separated the switched and unswitched B cells (going forward correspondingly referred to as memory and naïve B cells) from the individual donors with FACS and performed RNA extraction. The extracted RNA was depleted from ribosomal RNA, reverse transcribed, libraries generated and sent for 100bp single-end RNA sequencing using the Illumina Novaseq 6000 platform.
Results
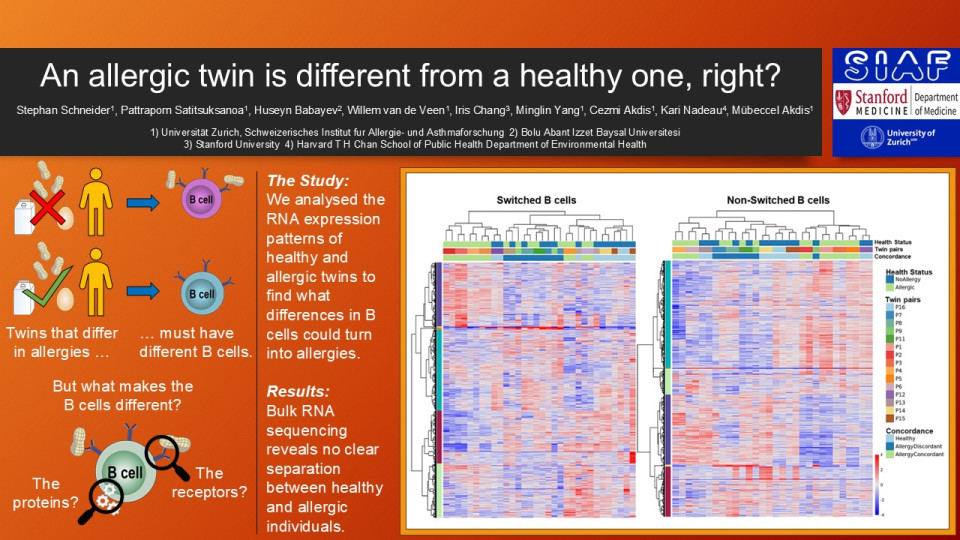
The unbiased clustering, without long non-coding RNA, indicates the grouping of the data points is determined by memory vs naïve B cells, twin pairs, and then by concordance status. Allergies had little to no weight for the clustering algorithm. PCA analysis of the top 300 significant genes show no clustering by health status as well. We further compared the healthy vs allergic donors of the discordant twin pairs gene expression analysis. Using a log fold change >0.5 as a base, we discovered that the FDR stayed above 0.99 until we reached a p-value threshold of <0.00001. This strongly indicates that any differences are purely random variation and not differently regulated B cell pathways across the whole B cell population.
The PCA analysis from discordant twins gives the same results with no clustering determined by allergy status of the donors.
Conclusion
We found no conclusive evidence that the underlying cause for allergies is a general dysregulation of B cells. In allergy discordant twin pairs, neither memory nor naïve B cells show overarching differences between healthy or allergic donors. Comparing the B cells between healthy and allergic twin pairs showed that the expression patterns are not as distinct as expected. What differences there are, are more likely contributed by differences between twin pairs than in allergy status. These results are counterintuitive to the perceived picture of allergies. Rather than a systemic difference in immune regulation, we propose that the effects are limited to the allergen specific B cells. As such we hypothesize that only allergen specific cells differ in their gene expression between allergic and healthy individuals. These effects most likely are overshadowed by overall variability due to how rare specific B cells are.