Background
Eosinophilic esophagitis (EoE) is a chronic, immune-mediated inflammatory condition that has shown increased prevalence in recent decades. The esophagus of affected patients is characterized by rings and furrows and severe cases can result in esophageal narrowing. The esophageal barrier serves as the primary site of inflammation, marked by the recruitment of more than 15 eosinophils per high power field (eos/hpf), in an area physiologically devoid of these cells. Diagnosing and monitoring EoE currently require invasive endoscopies due to the lack of non-invasive biomarkers.
Aim
Our objective is to identify plasma biomarkers capable of distinguishing between active and inactive EoE states to facilitate disease monitoring.
Method
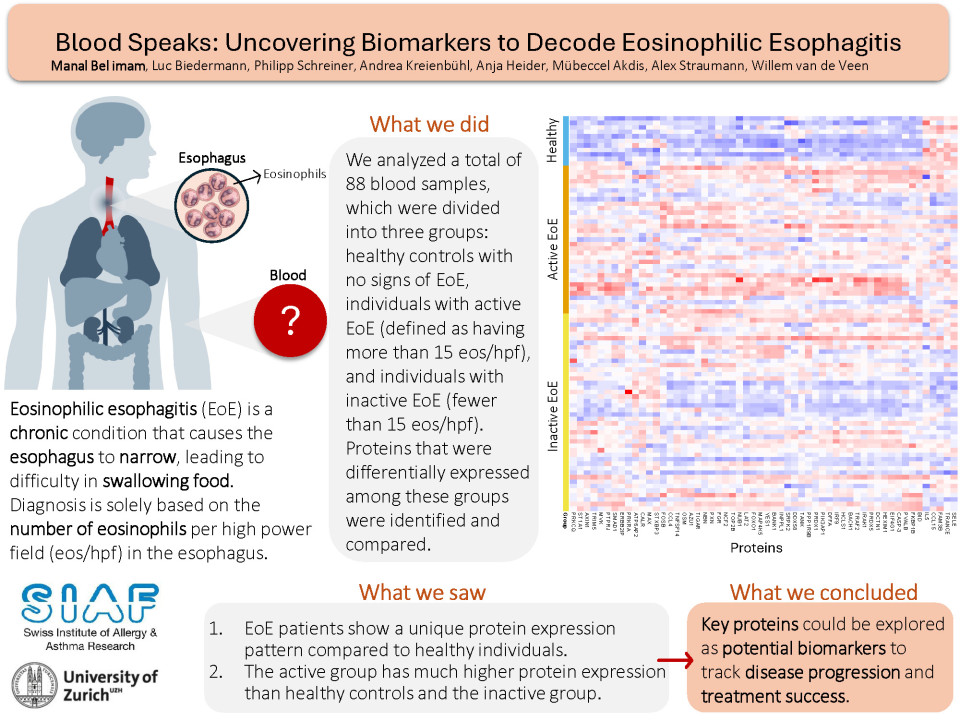
We collected blood samples from 142 EoE patients at the University Hospital Zurich and categorized them into active EoE (>15 eos/hpf) and inactive EoE (<15 eos/hpf). Using the Olink biomarker discovery system, we analysed the expression of 360 proteins in plasma samples from 77 EoE patients and 11 healthy controls.
Results
We identified 56 proteins that were differentially expressed in the groups (active EoE, inactive EoE and healthy controls). Among these, 17 proteins were significantly higher in the active EoE subgroup, including CCL4, TRANCE, and TRAF2. Only 1 protein, ATP6AP2, was higher in the inactive EoE subgroup compared to the active one.
Conclusions
Our analysis identified potential plasma biomarkers in EoE patients. To validate these findings, we will perform a targeted analysis of the proteins in a confirmation cohort. Additionally, we will assess their presence in the esophageal biopsies of the same patients to clarify their origins and better understand the mechanisms underlying the disease. Our research will provide valuable insights into EoE pathophysiology, while improving patients’ lives by reducing the number of endoscopies.