Eosinophilic esophagitis (EoE) is a Th2-mediated chronic inflammatory disorder, whose main hallmark is the recruitment of eosinophils into the esophagus, resulting in chronic inflammation, difficult swallowing, food impaction, and esophageal dysfunction. The prevalence of EoE has increased significantly worldwide, drawing more attention to this disease in the recent years.
Currently, EoE’s diagnosis and disease monitoring are based on histopathologic analyses which require repeated endoscopies and biopsies, representing expensive and invasive approaches for the patients.
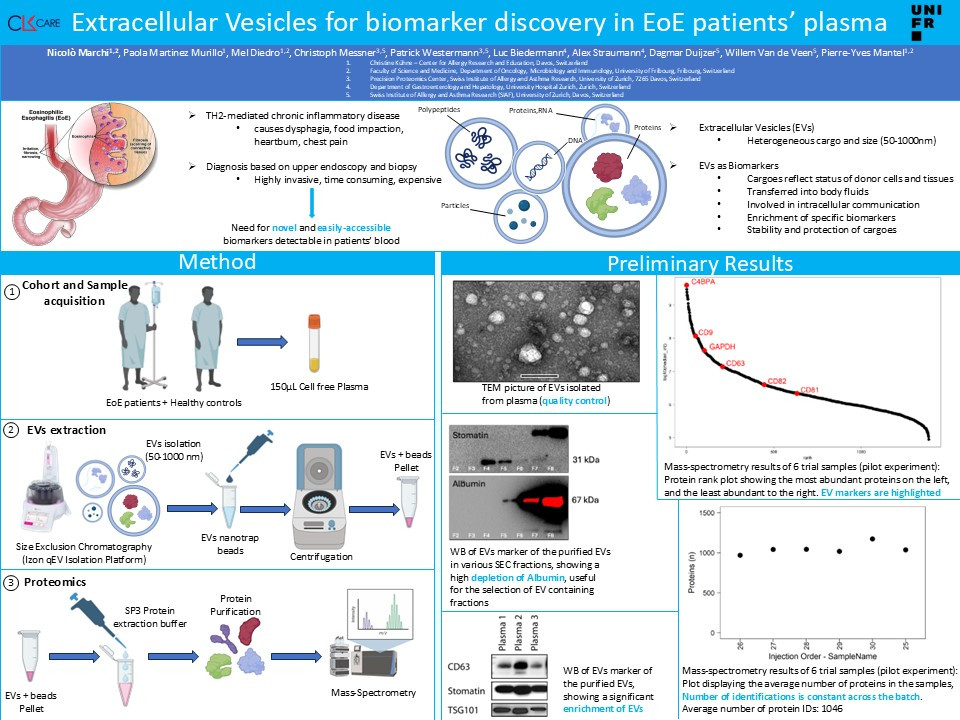
Therefore, there is an urgent need for novel technologies based on easily accessible biomarkers, that can be detected in the patients’ blood.
Compelling evidence has demonstrated the central role of epithelial barrier dysfunction in the pathophysiology of EoE, both in skewing the immune system towards allergic inflammation and in enabling the penetration of relevant antigens into the esophageal mucosa, leading to the above-mentioned symptoms. Moreover, numbers of inflammatory cells are increased in the affected epithelial space. Several single-cell RNA-Seq studies have further confirmed that epithelial cells, eosinophils, T lymphocytes (in particular a subtype of Th2 cells), mast cells, B lymphocytes and plasma cells are found in the lesions and might participate to the development of the disease. Interestingly, all these activated cells have the potential to secrete extracellular vesicles (EVs) that might be then found in the blood circulation.
EVs are a heterogeneous group of small vesicles involved in a great variety of physiological and pathological functions, which are released in the blood circulation by every cell type in the body and contain many different molecular cargoes derived from their originator cells. For this reason, there is growing attention to use them as biomarkers for difficult-to-diagnose cancers, such as pancreatic, prostate, or glioblastoma.
We are developing and validating a protocol to isolate EVs from plasma based on size exclusion chromatography (SEC). The quality of the isolated-EVs is validated by Transmission Electron Microscopy and Western Blotting to check for the presence of typical EV-markers and plasma protein contaminants. Here, we propose to characterize EVs in EoE patients’ plasma by Mass-spec based proteomics. We will use a cohort of about 100 patients for our discovery cohort. We will compare the EVs from this cohort with healthy controls. With this data, we will build a prediction model based on machine learning to identify a protein signature that identifies the disease. Our predictive model will then be validated by using a validation cohort, made of different patients than the ones included in the discovery cohort.