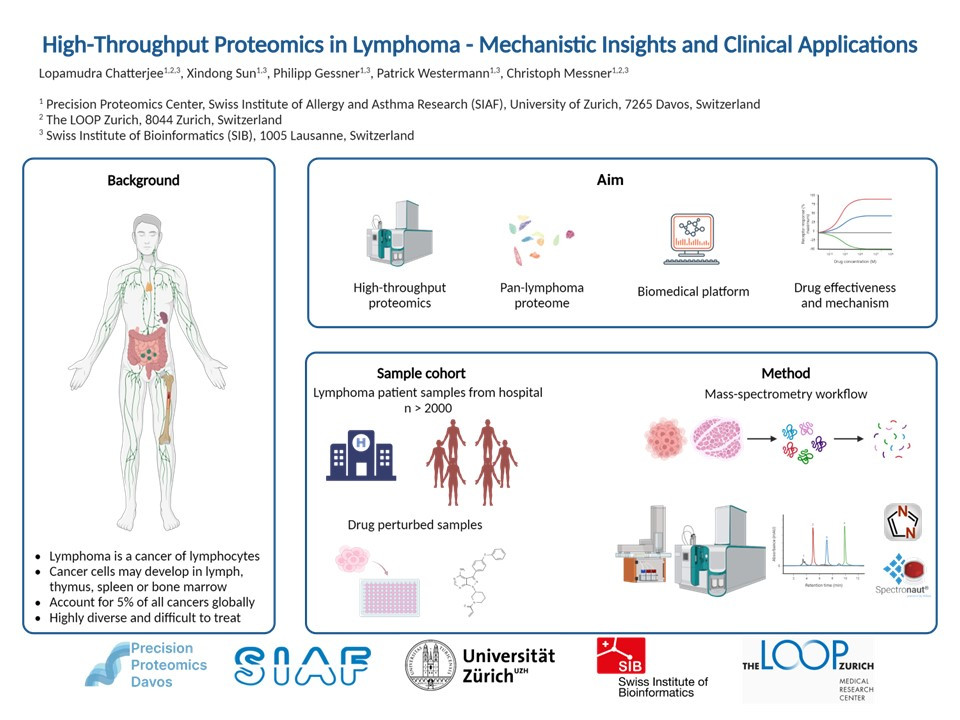
Lymphoma is a term for cancers that start in the cells of lymph system. Lymphomas account for 5% of all cancers globally and are a significant concern in the spectrum of hematologic diseases. Non Hodgkin Lymphoma (NHL) makes up the majority of lymphoma cases, accounting for approximately 85-90%, while Hodgkin lymphoma is rarer. Among different NHL types, B-cell NHL are more prevalent than T-cell NHLs. Over the last decade, genome based research have identified multiple types and subtypes of lymphomas. However, there are still several key questions about the onset and progression of the disease that remain unanswered. Lymphoma diagnosis, which relies mostly on immunophenotype and morphology, is challenging even for experts because of the disease's intricate mutational landscape. Another major concern in the field of lymphomas is that a significant number of patients cannot be treated with the current treatment methods that include: chemotherapy, radiotherapy, antibodies, kinase inhibitors and Chimeric Antigen Receptor T-cell (CAR-T cell).
Proteomics plays a crucial role in understanding disease characteristics, as alterations in protein expression, modification, and interaction are fundamental to the development and progression of diseases. However, proteomic analysis of lymphomas has been limited thus far primarily due to the technical challenges associated with it.
At the Precision Proteomics Center Davos, we currently have a very robust and sensitive mass spectrometry based approach that can process thousands of patient samples in a month. Our platform is compatible with different sample types like cells, plasma, serum as well as fresh-frozen and formalin fixed paraffin embedded tissue materials. Additionally, to aid with clinical translation, we are currently developing absolute quantification assays for different disease biomarkers, starting with lymphoma. With our technology, we want to generate the largest known proteomic repository of lymphoid malignancies.
The platform will serve as a proteome database encompassing the broadest possible range of B- and T-cell lymphomas. We expect that the comprehensive proteomic information along with molecularly annotated clinical data, will simplify the diagnosis of various lymphoma types and subtypes. Our current focus is mainly on more common lymphoma types like Chronic Lymphocytic Leukemia (CLL), Follicular Lymphoma (FL), Mantle Cell Lymphoma (MCL) and Diffuse Large B-Cell Lymphoma (DLBCL). In the later stages, we will then move onto diagnostically challenging and rarer lymphomas like T cell-Non Hodgkin Lymphoma (T-NHL) and Marginal Zone Lymphoma (MZL). The large-scale analysis of lymphomas using our robust proteomic platform should also enable the application of the database in clinical settings and potentially for routine diagnostics.
To further support clinical diagnostics, we are also conducting mass-spectrometry based drug perturbation assays. Proteomics can potentially measure the molecularly defined targets that the drugs are dependent upon. In the recent times, mass spectrometry has become the most comprehensive approach for proteome-wide characterization of drugs. It can be used to study both effectiveness of drugs and its underlying mechanism of action. These studies can also reveal biomarkers associated with drug response and resistance.
We are currently investigating how the proteome changes over time in response to drug treatment. Our aim is to identify early response biomarkers that predict treatment efficacy or resistance by comparing patient proteomic profiles at different time points after treatment.
We believe that using our expertise in high throughput proteomics and bioinformatics we will be able improve the diagnostic landscape of lymphomas, paving the way for personalized medicine and better treatment outcomes.