The medical coding of patient diagnoses and therapies in hospitals and other institutions plays a crucial role in the Swiss healthcare system. By converting clinical information into standardized codes, they enable more efficient data analyses, resource allocation and evidence-based policymaking. The usual medical coding process in Switzerland involves transforming clinical notes by doctors or other medical personnel, often only available in unstructured free-text format, into structured codes using classification systems like the “International Classification of Diseases – German Modification” (ICD-10-GM) for diagnoses and the “Schweizerische Operationsklassifikation (CHOP)” for treatments. At the national level, coded health data allows for epidemiological studies, monitoring health inequalities and strategic allocation of resources. For individual healthcare providers, like hospitals, these accurate clinical data also offer several optimization possibilities through efficient cost auditing, patient segmentation and patient journey mapping, decision support system development and ensuring appropriate reimbursement for services rendered. Currently many hospitals in Switzerland struggle to find enough personnel for the necessary medical coding work. To further support their work, automated clinical coding systems are being developed to further enhance efficiency and accuracy, addressing challenges such as handling long or incomplete medical records. Due to their performance and potential, Large Language Models (LLM) such as ChatGPT have emerged over the last few years as a disruptive technology across various research fields. Utilizing their architecture on huge amounts of unstructured text data, LLMs can emulate a deep understanding of language semantics. In their functionality, LLMs calculate the conditional probability of word sequences by taking into account the contextual information from foregoing words, so they can predict the probability of subsequent words. With their large context windows for inputs, their utilization requires fewer tedious tasks like chunking in comparison to traditional methods. As well, due to abundantly available extensive high-level libraries for working with LLMs, e.g. the Hugging-Face Transformers library, scripts are more accessible to a non-technical audience and less prone to error in their functionality implying more medical staff can make use of these scripts. The ease of LLM scripts also means they are easier to reproduce, making research more comprehensible and replicable.
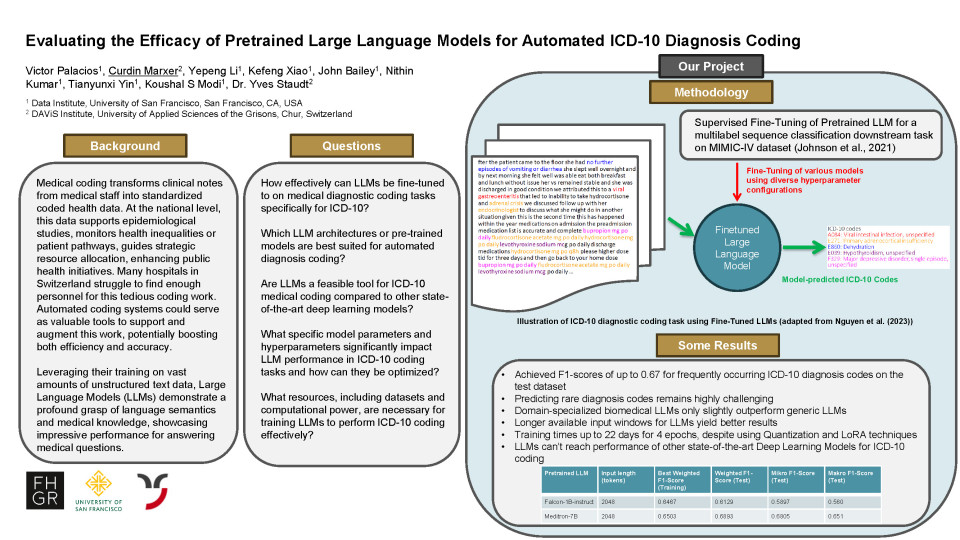
In our research we evaluate and benchmark the capabilities of different publicly available Large Language Models for automated ICD-10 coding of diagnoses. Using multilabel sequence classification as a downstream task for matching text sequences to their respective corresponding ICD-10 codes, we used the publicly available MIMIC-IV dataset to fine-tune cross-domain and domain-specialized biomedical LLMs spanning from 1 billion to 7 billion parameters. To optimize and reduce the necessary computing time, we employed multiple parameter-efficient fine-tuning (PEFT) methods to reduce the number of parameters of the LLMs, while maintaining satisfactory performance. One such prominent methods we employed is “Low-Rank Adaptation” (LoRA), which reduces the dimensionality of parameter matrices needed for computation. With another technique called “Quantization” we reduced the necessary GPU-bound memory space and the necessary number of calculations by converting the data representation of the weights and activations within an LLM from high to low-precision, e.g. a 32-bit floating-point number to an 8-bit integer.
For both the fine-tuning and the model evaluation process, the MIMIC-IV dataset provides more than 100’000 clinical notes of individual patients matched together with all corresponding ICD-10 diagnosis codes. Within our results we benchmark multiple LLMs with different parameters-sizes by comparing their F1-scores and observed necessary compute times for our defined downstream task. Our results show that even small cross-domain models like “Falcon-1B” achieve surprising results in predicting the corresponding ICD-10 codes for previously unknown text sequences.
Quiz question
When adapting pre-trained Large Language Models (Falcon-1B, Meditron-7B, and Phi-3.5B) for medical diagnosis coding (ICD-10), which of their existing features showed the most potential for improving our test results, despite overall performance challenges?
The amount of text they can process at once.
The number of model parameters they have.
Whether they were initially trained on medical or general information.