Introduction
Mouse models have become essential tools in studying bone healing and the underlying mechanisms of impaired healing and fracture non-union. These models replicate the clinical condition where fractured bones fail to heal properly, providing a controlled environment to investigate factors that contribute to healing impairments. Researchers utilize these models to test new treatments and understand the molecular and cellular processes involved in bone repair. In this context, we apply spatial transcriptomics – a cutting-edge technique that enables the visualization and spatial quantification of gene expression. By combining this technique with non-union mouse models, we gain a comprehensive understanding of the spatial and temporal dynamics of gene expression during bone healing.
Furthermore, the application of spatial transcriptomics extends to the evaluation of (calcium phosphate) bone graft substitutes, which are commonly used to promote bone regeneration. These substitutes provide a scaffold that supports new bone growth, and spatial transcriptomics can help elucidate the interactions between the graft materials and the host tissue at a molecular level.
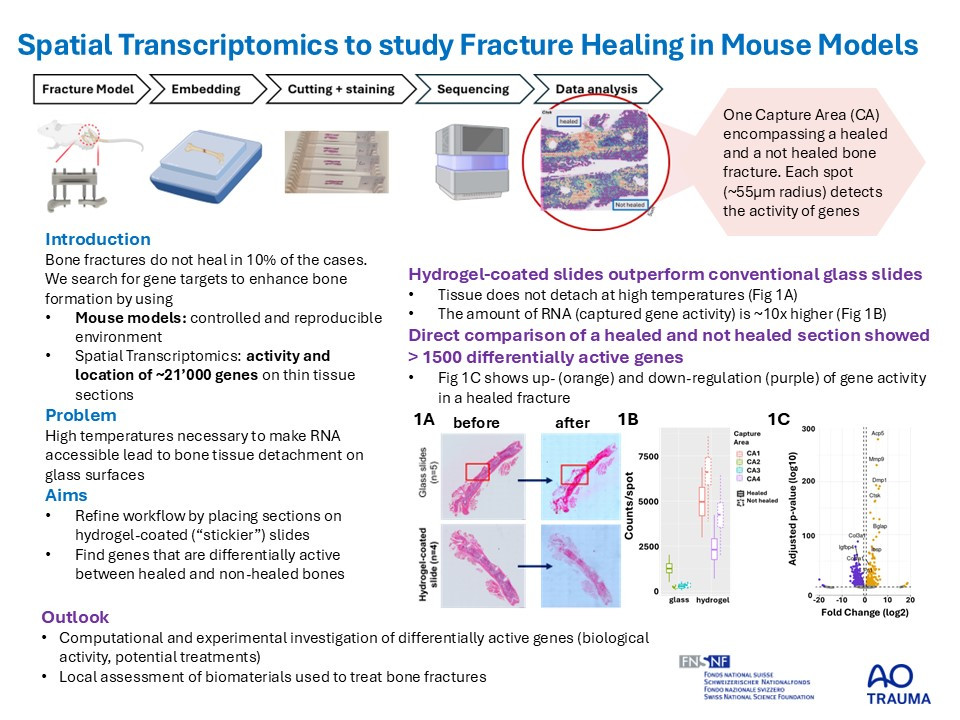
Refined spatial transcriptomics workflows based on transcriptomic probe transfer, such as the Visium CytAssist from 10x Genomics, promise to enhance these protocols by allowing researchers to pre-select tissue sections for more targeted analysis. However, the use of formalin-fixed paraffin-embedded (FFPE) musculoskeletal sections has posed challenges including tissue detachment during processing and a decline in RNA quality, which can affect the accuracy and reliability of the transcriptomic data obtained from these samples. Here we test the applicability of CytAssist on musculoskeletal tissues using conventional as well as hydrogel-coated histology slides. Subsequently, we investigate differences in data quality between sections on both slide types and assess differentially expressed genes (DEGs) between unions and non-unions.
Methods
Histological sections (n=8) used for spatial transcriptomics (Visium CytAssist FFPE; 10x Genomics, n=4 on glass sildes, n=4 on hydrogel-coated slides) were obtained from a fracture healing study in female 20-week-old C57BL/6J mice receiving either a femur osteotomy (0.7mm) or a segmental defect (2.4mm) (license 22/2022, Grisons CH). Musculoskeletal sections were placed on glass and hydrogel-coated slides, H&E stained, imaged, destained and decrosslinked according to CytAssist protocol with different decrosslinking temperature and section thickness. Sequence alignment was performed using SpaceRanger. Differential gene expression was performed using DESeq2 (Seurat) followed by Gene-Set-Enrichment-Analysis (GSEA) of Gene Ontology (ClusterProfiler). Group comparison of quality measures was done using a Welch’s t-test. Results are given as mean±standard deviation.
Results
In contrast to conventional glass slides, sections on hydrogel-coated slides stayed intact at high decrosslinking temperatures. Section thickness did not influence tissue detachment. We therefore sequenced 5µm sections using glass slides (n=4, decrosslinking temperature=70°C) and hydrogel-coated slides (n=4, decrosslinking temperature=95°C). Mean counts and genes per spot, were significantly ~10x higher for sections on hydrogel slides (counts: 4697±1796, genes: 2389±1170) compared to glass slides (counts: 463±415, genes: 250±223). Direct comparison of a non-union and union section showed a total of 564 DEGs.
Conclusions
Optimized spatial transcriptomics workflows based on transcriptomic probe transfer enable for improved read depth in musculoskeletal tissue enabling the characterization of molecular features discriminating non-union and union bone fractures. Within a recently launched SNSF Sinergia project (SLIHI4BONE)I, this method will allow to map gene expression to investigate how calcium phosphate bone graft substitutes can improve bone healing. By mapping gene expression patterns around the application site, researchers can gain insights into how these materials influence bone healing processes, aiming to optimized composition of bone graft substitutes and improving clinical outcomes.
Acknowledgements: AO Foundation (AOTRAUMA), SNSF (PhD salary), ChatGTP (OpenAI, 2024) assisted to improve the readability of the abstract.